stackstacxarray
When you use stackstac, you do not need to write any code to handle merging operations for many raster files. The data come out the other side in a neatly packaged xarray object with x, y, time, and band dimensions! This is very convenient when you are interested in an arbitrary area that may require combining data from many STAC items. The stackstac documentationstackstac + xarray workflow.
Define search parameters
I am interested in getting a mosaic of daily observations from Sentinel 2 for September 2022.
import pyprojimport pystac_clientimport stackstacimport xarray as xrfrom shapely.geometry import boxfrom shapely.ops import transform# STAC connection information for Sentinel 2 COGs = "https://earth-search.aws.element84.com/v1" = "sentinel-2-l2a" # spatial projection information = "epsg:5070" = pyproj.CRS.from_string(CRS_STRING).to_epsg()# area of interest along the North Shore of Lake Superior = box(373926 , 2744693 , 406338 , 2765304 )# a few more parameters = 100 # meters = ["red" , "green" , "blue" ]= "2022-09-01" = "2022-09-30"
Query the STAC for matching items
To query the STAC, we need to provide a bounding box in epsg:4326 coordinates:
# STAC items store bounding box info in epsg:4326 = pyproj.Transformer.from_crs(= CRS_STRING,= "epsg:4326" ,= True ,= transform(transformer_4326.transform, AOI).bounds
This will return all of the STAC items that intersect the provided bounding box and time window:
= pystac_client.Client.open (STAC_URL)= catalog.search(= [STAC_COLLECTION],= bbox_4326,= [START_DATE, END_DATE],
The query yields many STAC items, each of which describes multiple COGs (one per band). Using other tools, we would need to write a bunch of code to make sure we combine the data correctly but with stackstac we can forget about that and just get on with our analysis!
Stack it
Lazily load the raster data into an xarray.DataArray using stackstack.stack. This function uses the STAC item metadata to construct a multidimensional array with human-readable coordinates that can be manipulated with the magnificently powerful suite of xarray functions and methods!
= stackstac.stack(= stac_items,= BANDS,= EPSG,= RESOLUTION,= AOI.bounds,= "center" ,
<xarray.DataArray 'stackstac-5efc1f91909c10ae7d8453451c8ac811' (time: 20,
band: 3,
y: 208, x: 325)> Size: 32MB
dask.array<fetch_raster_window, shape=(20, 3, 208, 325), dtype=float64, chunksize=(1, 1, 208, 325), chunktype=numpy.ndarray>
Coordinates: (12/54)
* time (time) datetime64[ns] 160B 2022-...
id (time) <U24 2kB 'S2B_15UXP_20220...
* band (band) <U5 60B 'red' 'green' 'blue'
* x (x) float64 3kB 3.74e+05 ... 4.0...
* y (y) float64 2kB 2.765e+06 ... 2....
s2:nodata_pixel_percentage (time) object 160B 51.473683 ......
... ...
gsd int64 8B 10
raster:bands object 8B {'nodata': 0, 'data_ty...
common_name (band) <U5 60B 'red' 'green' 'blue'
center_wavelength (band) float64 24B 0.665 0.56 0.49
full_width_half_max (band) float64 24B 0.038 ... 0.098
epsg int64 8B 5070
Attributes:
spec: RasterSpec(epsg=5070, bounds=(373900, 2744600, 406400, 27654...
crs: epsg:5070
transform: | 100.00, 0.00, 373900.00|\n| 0.00,-100.00, 2765400.00|\n| 0...
resolution: 100 dask.array<chunksize=(1, 1, 208, 325), meta=np.ndarray>
Bytes
30.94 MiB
528.12 kiB
Shape
(20, 3, 208, 325)
(1, 1, 208, 325)
Dask graph
60 chunks in 3 graph layers
Data type
float64 numpy.ndarray
20 1 325 208 3
Coordinates: (54)
time
(time)
datetime64[ns]
2022-09-03T17:10:24.557000 ... 2...
array(['2022-09-03T17:10:24.557000000', '2022-09-03T17:10:34.747000000',
'2022-09-06T17:20:16.965000000', '2022-09-06T17:20:29.331000000',
'2022-09-08T17:10:30.113000000', '2022-09-08T17:10:44.490000000',
'2022-09-13T17:10:20.922000000', '2022-09-13T17:10:35.306000000',
'2022-09-16T17:20:17.041000000', '2022-09-16T17:20:30.820000000',
'2022-09-18T17:10:27.345000000', '2022-09-18T17:10:41.728000000',
'2022-09-21T17:20:23.086000000', '2022-09-21T17:20:36.246000000',
'2022-09-23T17:10:19.894000000', '2022-09-23T17:10:34.274000000',
'2022-09-26T17:20:22.190000000', '2022-09-26T17:20:29.371000000',
'2022-09-28T17:10:26.238000000', '2022-09-28T17:10:40.620000000'],
dtype='datetime64[ns]') id
(time)
<U24
'S2B_15UXP_20220903_0_L2A' ... '...
array(['S2B_15UXP_20220903_0_L2A', 'S2B_15TXN_20220903_0_L2A',
'S2B_15UXP_20220906_0_L2A', 'S2B_15TXN_20220906_1_L2A',
'S2A_15UXP_20220908_0_L2A', 'S2A_15TXN_20220908_0_L2A',
'S2B_15UXP_20220913_0_L2A', 'S2B_15TXN_20220913_0_L2A',
'S2B_15UXP_20220916_0_L2A', 'S2B_15TXN_20220916_0_L2A',
'S2A_15UXP_20220918_0_L2A', 'S2A_15TXN_20220918_0_L2A',
'S2A_15UXP_20220921_1_L2A', 'S2A_15TXN_20220921_0_L2A',
'S2B_15UXP_20220923_0_L2A', 'S2B_15TXN_20220923_0_L2A',
'S2B_15UXP_20220926_1_L2A', 'S2B_15TXN_20220926_1_L2A',
'S2A_15UXP_20220928_0_L2A', 'S2A_15TXN_20220928_0_L2A'],
dtype='<U24') band
(band)
<U5
'red' 'green' 'blue'
array(['red', 'green', 'blue'], dtype='<U5') x
(x)
float64
3.74e+05 3.74e+05 ... 4.064e+05
array([373950., 374050., 374150., ..., 406150., 406250., 406350.]) y
(y)
float64
2.765e+06 2.765e+06 ... 2.745e+06
array([2765350., 2765250., 2765150., ..., 2744850., 2744750., 2744650.]) s2:nodata_pixel_percentage
(time)
object
51.473683 0.040103 ... 3.152056
array([51.473683, 0.040103, 45.561698, 76.072884, 0, 0, 3e-06, 4e-05,
46.01481, 71.087068, 0, 0, 49.96967, 71.087909, 7e-06, 0,
87.015414, 71.081239, 0.072037, 3.152056], dtype=object) s2:cloud_shadow_percentage
(time)
object
0.4397 0.747305 ... 0 0.087879
array([0.4397, 0.747305, 6.811442, 10.38455, 0.397139, 0.169246, 0.661142,
0, 0.393296, 0.546843, 0.0863, 0, 8.853565, 2.392711, 0.842539,
0.056546, 3.4027, 4.373321, 0, 0.087879], dtype=object) s2:thin_cirrus_percentage
(time)
float64
2.422 5.615 0.01568 ... 14.32 12.02
array([2.4218710e+00, 5.6151000e+00, 1.5682000e-02, 1.7190000e-03,
3.3271290e+00, 2.6672861e+01, 8.2513160e+00, 3.0100340e+00,
1.3676500e+00, 4.0137880e+00, 6.7195200e-01, 5.2816300e-01,
2.3496000e-02, 6.5675000e-02, 1.8442127e+01, 1.0292700e+00,
2.3051340e+00, 3.2298450e+00, 1.4315028e+01, 1.2021706e+01]) s2:water_percentage
(time)
object
13.552162 49.923825 ... 50.608224
array([13.552162, 49.923825, 14.386694, 4.856194, 0.271326, 23.370194,
11.297255, 59.993666, 0.040372, 0.016513, 0.017857, 0, 7.190147,
5.991462, 2.222859, 0.257096, 3.026956, 3.399163, 12.471328,
50.608224], dtype=object) s2:unclassified_percentage
(time)
object
0.530771 0.126185 ... 0.010277
array([0.530771, 0.126185, 9.449986, 4.049694, 0.099296, 0.021689,
0.622211, 0.00135, 0, 0, 0.000385, 0, 5.935226, 3.792643, 0.068739,
0.000484, 1.850153, 2.082388, 0.000269, 0.010277], dtype=object) mgrs:utm_zone
()
int64
15
eo:cloud_cover
(time)
object
3.568175 13.802318 ... 12.126758
array([3.568175, 13.802318, 19.605383, 25.269425, 99.232119, 72.870868,
22.926284, 3.010038, 99.56134, 99.435449, 99.894994, 100, 60.79812,
77.379012, 79.006791, 99.408644, 85.670727, 67.162913, 14.315172,
12.126758], dtype=object) s2:vegetation_percentage
(time)
object
81.094176 35.154691 ... 36.676258
array([81.094176, 35.154691, 46.135363, 53.639263, 0, 3.169266, 53.416437,
36.590198, 0, 0, 0.000199, 0, 15.932353, 10.140032, 17.190969,
0.274326, 5.537937, 22.063114, 71.710277, 36.676258], dtype=object) s2:product_type
()
<U7
'S2MSI2A'
array('S2MSI2A', dtype='<U7') earthsearch:s3_path
(time)
<U79
's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2B_15UXP_20220903_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2B_15TXN_20220903_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2B_15UXP_20220906_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2B_15TXN_20220906_1_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2A_15UXP_20220908_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2A_15TXN_20220908_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2B_15UXP_20220913_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2B_15TXN_20220913_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2B_15UXP_20220916_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2B_15TXN_20220916_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2A_15UXP_20220918_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2A_15TXN_20220918_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2A_15UXP_20220921_1_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2A_15TXN_20220921_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2B_15UXP_20220923_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2B_15TXN_20220923_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2B_15UXP_20220926_1_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2B_15TXN_20220926_1_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/U/XP/2022/9/S2A_15UXP_20220928_0_L2A',
's3://sentinel-cogs/sentinel-s2-l2a-cogs/15/T/XN/2022/9/S2A_15TXN_20220928_0_L2A'],
dtype='<U79') s2:degraded_msi_data_percentage
(time)
object
0.0124 0.0023 0.0308 ... 0.0116 0 0
array([0.0124, 0.0023, 0.0308, 0.0149, 0, 0, 0.0107, 0.0021, 0.0307,
0.0116, 0, 0, 0.0245, 0, 0.0107, 0.002, 0.0278, 0.0116, 0, 0],
dtype=object) earthsearch:payload_id
(time)
<U74
'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/adc824c409650ba600ee2717785e90b4',
'roda-sentinel2/workflow-sentinel2-to-stac/92789b9ca5e71c603bb10d6f5300600f',
'roda-sentinel2/workflow-sentinel2-to-stac/4d9b7ca88d8696e21a7ebd2b1943fa53',
'roda-sentinel2/workflow-sentinel2-to-stac/07c9f3498fc327200d57b6d1e35af498',
'roda-sentinel2/workflow-sentinel2-to-stac/d2cc38087bfefc7a5372c8c34b62e91d',
'roda-sentinel2/workflow-sentinel2-to-stac/ced88ab580dbff0980fbf59ab09677c5',
'roda-sentinel2/workflow-sentinel2-to-stac/7058ba98237cc5b94d48c0cdea0f5c8c',
'roda-sentinel2/workflow-sentinel2-to-stac/0f68385cd38faced11118f802afc6d85',
'roda-sentinel2/workflow-sentinel2-to-stac/47badc0d06a3a6bc08c3ec3b5f8e21e5',
'roda-sentinel2/workflow-sentinel2-to-stac/4f6f42097f409e46003f52b5bf492cb2',
'roda-sentinel2/workflow-sentinel2-to-stac/a998072eb97b895123c0a40fcb99cbbb',
'roda-sentinel2/workflow-sentinel2-to-stac/a7c31b8628b4308109f4dde0e8250c5f',
'roda-sentinel2/workflow-sentinel2-to-stac/4dbefc9f362f9ac4c96f71def77a8eb6',
'roda-sentinel2/workflow-sentinel2-to-stac/9fb4aa81005abf9387602f3f8e513458',
'roda-sentinel2/workflow-sentinel2-to-stac/0b9db3ce1c76563c4941b170a42ebeec',
'roda-sentinel2/workflow-sentinel2-to-stac/16cf1664f192ed8100f602dbfcc22ff7',
'roda-sentinel2/workflow-sentinel2-to-stac/b0ae933f21e101e6043dc33c9a56a2fb',
'roda-sentinel2/workflow-sentinel2-to-stac/0d325815422d36ca93c9c025dea4c991',
'roda-sentinel2/workflow-sentinel2-to-stac/17787e79fba62312bd72ecb91d2e14c2',
'roda-sentinel2/workflow-sentinel2-to-stac/3b443d19641dea8a61cc6ca72de8b42e'],
dtype='<U74') s2:dark_features_percentage
(time)
object
0.139082 0.00942 0.864926 ... 0 0 0
array([0.139082, 0.00942, 0.864926, 0.038937, 0, 0.012886, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=object) s2:not_vegetated_percentage
(time)
object
0.675932 0.236259 ... 0.490598
array([0.675932, 0.236259, 2.746207, 1.761939, 4.6e-05, 0.385851,
11.076669, 0.404747, 0, 5.7e-05, 0, 0, 1.290586, 0.304127,
0.668107, 0.002903, 0.511528, 0.919099, 1.502949, 0.490598],
dtype=object) updated
(time)
<U24
'2022-11-06T12:04:29.428Z' ... '...
array(['2022-11-06T12:04:29.428Z', '2022-11-06T13:12:39.489Z',
'2022-11-05T22:16:39.665Z', '2022-11-06T12:55:37.142Z',
'2022-11-06T12:04:38.178Z', '2022-11-06T12:55:06.934Z',
'2022-11-06T12:04:32.450Z', '2022-11-06T12:54:01.620Z',
'2022-11-06T12:07:29.036Z', '2022-11-05T22:48:38.559Z',
'2022-11-06T12:07:31.465Z', '2022-11-06T12:55:18.668Z',
'2022-11-06T12:27:21.452Z', '2022-11-06T12:56:04.305Z',
'2022-11-06T12:23:48.863Z', '2022-11-06T13:18:48.730Z',
'2022-11-06T12:04:23.440Z', '2022-11-06T12:54:42.487Z',
'2022-11-06T12:04:28.328Z', '2022-11-06T13:12:39.171Z'],
dtype='<U24') s2:reflectance_conversion_factor
(time)
float64
0.9816 0.9816 ... 0.9946 0.9946
array([0.98156088, 0.98156088, 0.98298171, 0.98298171, 0.98395027,
0.98395027, 0.9864659 , 0.9864659 , 0.98803248, 0.98803248,
0.98909105, 0.98909105, 0.99071468, 0.99071468, 0.99180686,
0.99180686, 0.99347586, 0.99347586, 0.99459435, 0.99459435]) s2:processing_baseline
()
<U5
'04.00'
array('04.00', dtype='<U5') instruments
()
<U3
'msi'
array('msi', dtype='<U3') s2:datatake_type
()
<U8
'INS-NOBS'
array('INS-NOBS', dtype='<U8') s2:generation_time
(time)
<U27
'2022-09-03T21:41:48.000000Z' .....
array(['2022-09-03T21:41:48.000000Z', '2022-09-03T21:41:48.000000Z',
'2022-09-06T21:05:22.000000Z', '2022-09-06T21:05:22.000000Z',
'2022-09-09T01:07:53.000000Z', '2022-09-09T01:07:53.000000Z',
'2022-09-14T06:32:24.000000Z', '2022-09-14T06:32:24.000000Z',
'2022-09-16T21:37:47.000000Z', '2022-09-16T21:37:47.000000Z',
'2022-09-18T23:06:59.000000Z', '2022-09-18T23:06:59.000000Z',
'2022-09-21T23:09:55.000000Z', '2022-09-21T23:09:55.000000Z',
'2022-09-24T17:20:45.000000Z', '2022-09-24T17:20:45.000000Z',
'2022-09-26T21:25:54.000000Z', '2022-09-26T21:25:54.000000Z',
'2022-09-28T23:02:58.000000Z', '2022-09-28T23:02:58.000000Z'],
dtype='<U27') s2:granule_id
(time)
<U62
'S2B_OPER_MSI_L2A_TL_2BPS_202209...
array(['S2B_OPER_MSI_L2A_TL_2BPS_20220903T214148_A028695_T15UXP_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220903T214148_A028695_T15TXN_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220906T210522_A028738_T15UXP_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220906T210522_A028738_T15TXN_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220909T010753_A037675_T15UXP_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220909T010753_A037675_T15TXN_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220914T063224_A028838_T15UXP_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220914T063224_A028838_T15TXN_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220916T213747_A028881_T15UXP_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220916T213747_A028881_T15TXN_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220918T230659_A037818_T15UXP_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220918T230659_A037818_T15TXN_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220921T230955_A037861_T15UXP_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220921T230955_A037861_T15TXN_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220924T172045_A028981_T15UXP_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220924T172045_A028981_T15TXN_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220926T212554_A029024_T15UXP_N04.00',
'S2B_OPER_MSI_L2A_TL_2BPS_20220926T212554_A029024_T15TXN_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220928T230258_A037961_T15UXP_N04.00',
'S2A_OPER_MSI_L2A_TL_ATOS_20220928T230258_A037961_T15TXN_N04.00'],
dtype='<U62') mgrs:latitude_band
(time)
<U1
'U' 'T' 'U' 'T' ... 'U' 'T' 'U' 'T'
array(['U', 'T', 'U', 'T', 'U', 'T', 'U', 'T', 'U', 'T', 'U', 'T', 'U',
'T', 'U', 'T', 'U', 'T', 'U', 'T'], dtype='<U1') mgrs:grid_square
(time)
<U2
'XP' 'XN' 'XP' ... 'XN' 'XP' 'XN'
array(['XP', 'XN', 'XP', 'XN', 'XP', 'XN', 'XP', 'XN', 'XP', 'XN', 'XP',
'XN', 'XP', 'XN', 'XP', 'XN', 'XP', 'XN', 'XP', 'XN'], dtype='<U2') view:sun_azimuth
(time)
float64
160.4 160.1 164.7 ... 165.9 165.8
array([160.43367982, 160.13283535, 164.70429313, 164.47937286,
161.68231494, 161.41986299, 162.78628849, 162.5573591 ,
166.82512759, 166.66062771, 163.92909871, 163.73144666,
167.8380842 , 167.69924537, 164.93165357, 164.76087546,
168.7157714 , 168.59851679, 165.93563454, 165.78945588]) s2:datatake_id
(time)
<U34
'GS2B_20220903T165849_028695_N04...
array(['GS2B_20220903T165849_028695_N04.00',
'GS2B_20220903T165849_028695_N04.00',
'GS2B_20220906T170849_028738_N04.00',
'GS2B_20220906T170849_028738_N04.00',
'GS2A_20220908T165901_037675_N04.00',
'GS2A_20220908T165901_037675_N04.00',
'GS2B_20220913T165849_028838_N04.00',
'GS2B_20220913T165849_028838_N04.00',
'GS2B_20220916T170849_028881_N04.00',
'GS2B_20220916T170849_028881_N04.00',
'GS2A_20220918T165951_037818_N04.00',
'GS2A_20220918T165951_037818_N04.00',
'GS2A_20220921T171021_037861_N04.00',
'GS2A_20220921T171021_037861_N04.00',
'GS2B_20220923T170029_028981_N04.00',
'GS2B_20220923T170029_028981_N04.00',
'GS2B_20220926T171049_029024_N04.00',
'GS2B_20220926T171049_029024_N04.00',
'GS2A_20220928T170111_037961_N04.00',
'GS2A_20220928T170111_037961_N04.00'], dtype='<U34') s2:product_uri
(time)
<U65
'S2B_MSIL2A_20220903T165849_N040...
array(['S2B_MSIL2A_20220903T165849_N0400_R069_T15UXP_20220903T214148.SAFE',
'S2B_MSIL2A_20220903T165849_N0400_R069_T15TXN_20220903T214148.SAFE',
'S2B_MSIL2A_20220906T170849_N0400_R112_T15UXP_20220906T210522.SAFE',
'S2B_MSIL2A_20220906T170849_N0400_R112_T15TXN_20220906T210522.SAFE',
'S2A_MSIL2A_20220908T165901_N0400_R069_T15UXP_20220909T010753.SAFE',
'S2A_MSIL2A_20220908T165901_N0400_R069_T15TXN_20220909T010753.SAFE',
'S2B_MSIL2A_20220913T165849_N0400_R069_T15UXP_20220914T063224.SAFE',
'S2B_MSIL2A_20220913T165849_N0400_R069_T15TXN_20220914T063224.SAFE',
'S2B_MSIL2A_20220916T170849_N0400_R112_T15UXP_20220916T213747.SAFE',
'S2B_MSIL2A_20220916T170849_N0400_R112_T15TXN_20220916T213747.SAFE',
'S2A_MSIL2A_20220918T165951_N0400_R069_T15UXP_20220918T230659.SAFE',
'S2A_MSIL2A_20220918T165951_N0400_R069_T15TXN_20220918T230659.SAFE',
'S2A_MSIL2A_20220921T171021_N0400_R112_T15UXP_20220921T230955.SAFE',
'S2A_MSIL2A_20220921T171021_N0400_R112_T15TXN_20220921T230955.SAFE',
'S2B_MSIL2A_20220923T170029_N0400_R069_T15UXP_20220924T172045.SAFE',
'S2B_MSIL2A_20220923T170029_N0400_R069_T15TXN_20220924T172045.SAFE',
'S2B_MSIL2A_20220926T171049_N0400_R112_T15UXP_20220926T212554.SAFE',
'S2B_MSIL2A_20220926T171049_N0400_R112_T15TXN_20220926T212554.SAFE',
'S2A_MSIL2A_20220928T170111_N0400_R069_T15UXP_20220928T230258.SAFE',
'S2A_MSIL2A_20220928T170111_N0400_R069_T15TXN_20220928T230258.SAFE'],
dtype='<U65') s2:saturated_defective_pixel_percentage
()
int64
0
s2:high_proba_clouds_percentage
(time)
object
0.05344 3.811854 ... 7e-06 0.037061
array([0.05344, 3.811854, 8.700146, 11.212917, 82.789242, 34.899801,
10.065806, 0, 1.227298, 13.973652, 95.872045, 69.376743, 46.87703,
64.019388, 33.67047, 48.321438, 73.373586, 51.590377, 7e-06,
0.037061], dtype=object) s2:medium_proba_clouds_percentage
(time)
float64
1.093 4.375 ... 0.000136 0.06799
array([1.0928640e+00, 4.3753640e+00, 1.0889556e+01, 1.4054789e+01,
1.3115746e+01, 1.1298204e+01, 4.6091620e+00, 3.0000000e-06,
9.6966392e+01, 8.1448013e+01, 3.3510010e+00, 3.0095091e+01,
1.3897596e+01, 1.3293955e+01, 2.6894188e+01, 5.0057936e+01,
9.9920050e+00, 1.2342693e+01, 1.3600000e-04, 6.7992000e-02]) created
(time)
<U24
'2022-11-06T12:04:29.428Z' ... '...
array(['2022-11-06T12:04:29.428Z', '2022-11-06T13:12:39.489Z',
'2022-11-05T22:16:39.665Z', '2022-11-06T12:55:37.142Z',
'2022-11-06T12:04:38.178Z', '2022-11-06T12:55:06.934Z',
'2022-11-06T12:04:32.450Z', '2022-11-06T12:54:01.620Z',
'2022-11-06T12:07:29.036Z', '2022-11-05T22:48:38.559Z',
'2022-11-06T12:07:31.465Z', '2022-11-06T12:55:18.668Z',
'2022-11-06T12:27:21.452Z', '2022-11-06T12:56:04.305Z',
'2022-11-03T07:22:49.467Z', '2022-11-06T13:18:48.730Z',
'2022-11-03T07:13:04.965Z', '2022-11-06T12:54:42.487Z',
'2022-11-06T12:04:28.328Z', '2022-11-06T13:12:39.171Z'],
dtype='<U24') earthsearch:boa_offset_applied
(time)
bool
True True True ... True True True
array([ True, True, True, True, True, True, True, True, True,
True, True, True, True, True, False, True, False, True,
True, True]) processing:software
()
object
{'sentinel2-to-stac': '0.1.0'}
array({'sentinel2-to-stac': '0.1.0'}, dtype=object) s2:sequence
(time)
<U1
'0' '0' '0' '1' ... '1' '1' '0' '0'
array(['0', '0', '0', '1', '0', '0', '0', '0', '0', '0', '0', '0', '1',
'0', '0', '0', '1', '1', '0', '0'], dtype='<U1') proj:epsg
()
int64
32615
grid:code
(time)
<U10
'MGRS-15UXP' ... 'MGRS-15TXN'
array(['MGRS-15UXP', 'MGRS-15TXN', 'MGRS-15UXP', 'MGRS-15TXN',
'MGRS-15UXP', 'MGRS-15TXN', 'MGRS-15UXP', 'MGRS-15TXN',
'MGRS-15UXP', 'MGRS-15TXN', 'MGRS-15UXP', 'MGRS-15TXN',
'MGRS-15UXP', 'MGRS-15TXN', 'MGRS-15UXP', 'MGRS-15TXN',
'MGRS-15UXP', 'MGRS-15TXN', 'MGRS-15UXP', 'MGRS-15TXN'],
dtype='<U10') s2:snow_ice_percentage
(time)
object
0 0 0 0 7.3e-05 0 0 ... 0 0 0 0 0 0
array([0, 0, 0, 0, 7.3e-05, 0, 0, 0, 0.00499, 0.001136, 0.000262, 0, 0,
1.1e-05, 0, 0, 0, 0, 0, 0], dtype=object) platform
(time)
<U11
'sentinel-2b' ... 'sentinel-2a'
array(['sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b',
'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b',
'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a',
'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b',
'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a'],
dtype='<U11') s2:datastrip_id
(time)
<U64
'S2B_OPER_MSI_L2A_DS_2BPS_202209...
array(['S2B_OPER_MSI_L2A_DS_2BPS_20220903T214148_S20220903T171016_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220903T214148_S20220903T171016_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220906T210522_S20220906T170849_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220906T210522_S20220906T170849_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220909T010753_S20220908T170317_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220909T010753_S20220908T170317_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220914T063224_S20220913T170241_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220914T063224_S20220913T170241_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220916T213747_S20220916T171756_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220916T213747_S20220916T171756_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220918T230659_S20220918T170147_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220918T230659_S20220918T170147_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220921T230955_S20220921T172012_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220921T230955_S20220921T172012_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220924T172045_S20220923T170540_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220924T172045_S20220923T170540_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220926T212554_S20220926T172015_N04.00',
'S2B_OPER_MSI_L2A_DS_2BPS_20220926T212554_S20220926T172015_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220928T230258_S20220928T170926_N04.00',
'S2A_OPER_MSI_L2A_DS_ATOS_20220928T230258_S20220928T170926_N04.00'],
dtype='<U64') view:sun_elevation
(time)
float64
47.63 48.48 47.09 ... 38.65 39.53
array([47.63026752, 48.48206693, 47.08586735, 47.95673495, 45.91375513,
46.77169169, 44.13629567, 44.99932078, 43.47927954, 44.3578936 ,
42.33530371, 43.20323453, 41.63041727, 42.51228789, 40.49702661,
41.36896547, 39.75615638, 40.64061823, 38.65330281, 39.52898369]) constellation
()
<U10
'sentinel-2'
array('sentinel-2', dtype='<U10') s2:mgrs_tile
(time)
object
None None '15UXP' ... None None
array([None, None, '15UXP', None, None, None, None, None, None, '15TXN',
None, None, None, None, None, None, None, None, None, None],
dtype=object) title
(band)
<U20
'Red (band 4) - 10m' ... 'Blue (...
array(['Red (band 4) - 10m', 'Green (band 3) - 10m',
'Blue (band 2) - 10m'], dtype='<U20') proj:shape
()
object
{10980}
array({10980}, dtype=object) gsd
()
int64
10
raster:bands
()
object
{'nodata': 0, 'data_type': 'uint...
array({'nodata': 0, 'data_type': 'uint16', 'bits_per_sample': 15, 'spatial_resolution': 10, 'scale': 0.0001, 'offset': -0.1},
dtype=object) common_name
(band)
<U5
'red' 'green' 'blue'
array(['red', 'green', 'blue'], dtype='<U5') center_wavelength
(band)
float64
0.665 0.56 0.49
array([0.665, 0.56 , 0.49 ]) full_width_half_max
(band)
float64
0.038 0.045 0.098
array([0.038, 0.045, 0.098]) epsg
()
int64
5070
Indexes: (4)
PandasIndex
PandasIndex(DatetimeIndex(['2022-09-03 17:10:24.557000', '2022-09-03 17:10:34.747000',
'2022-09-06 17:20:16.965000', '2022-09-06 17:20:29.331000',
'2022-09-08 17:10:30.113000', '2022-09-08 17:10:44.490000',
'2022-09-13 17:10:20.922000', '2022-09-13 17:10:35.306000',
'2022-09-16 17:20:17.041000', '2022-09-16 17:20:30.820000',
'2022-09-18 17:10:27.345000', '2022-09-18 17:10:41.728000',
'2022-09-21 17:20:23.086000', '2022-09-21 17:20:36.246000',
'2022-09-23 17:10:19.894000', '2022-09-23 17:10:34.274000',
'2022-09-26 17:20:22.190000', '2022-09-26 17:20:29.371000',
'2022-09-28 17:10:26.238000', '2022-09-28 17:10:40.620000'],
dtype='datetime64[ns]', name='time', freq=None)) PandasIndex
PandasIndex(Index(['red', 'green', 'blue'], dtype='object', name='band')) PandasIndex
PandasIndex(Index([373950.0, 374050.0, 374150.0, 374250.0, 374350.0, 374450.0, 374550.0,
374650.0, 374750.0, 374850.0,
...
405450.0, 405550.0, 405650.0, 405750.0, 405850.0, 405950.0, 406050.0,
406150.0, 406250.0, 406350.0],
dtype='float64', name='x', length=325)) PandasIndex
PandasIndex(Index([2765350.0, 2765250.0, 2765150.0, 2765050.0, 2764950.0, 2764850.0,
2764750.0, 2764650.0, 2764550.0, 2764450.0,
...
2745550.0, 2745450.0, 2745350.0, 2745250.0, 2745150.0, 2745050.0,
2744950.0, 2744850.0, 2744750.0, 2744650.0],
dtype='float64', name='y', length=208)) Attributes: (4)
spec : RasterSpec(epsg=5070, bounds=(373900, 2744600, 406400, 2765400), resolutions_xy=(100, 100)) crs : epsg:5070 transform : | 100.00, 0.00, 373900.00|
| 0.00,-100.00, 2765400.00|
| 0.00, 0.00, 1.00| resolution : 100
The resolution argument makes it possible to resample the input data on-the-fly. In this case, I am downsampling from the original 20 meter resolution to 100 meters.
Wrangle the time dimension
One thing to watch out for with stackstac.stack is that you will wind up with a distinct time coordinate for each STAC item that you pass in. To achieve the intuitive representation of the data, you need to flatten the DataArray with respect to day.
Note: if you are only reading a single STAC item, stackstac.mosaic will inadvertently reduce your data along the band dimension (which is definitely not what you want!), hence the conditional statement checking for more than one time coordinate value.
def flatten(x, dim= "time" ):assert isinstance (x, xr.DataArray)if len (x[dim].values) > len (set (x[dim].values)):= x.groupby(dim).map (stackstac.mosaic)return x# round time coordinates so all observations from the same day so they have # equivalent timestamps = sentinel_stack.assign_coords(= sentinel_stack.time.astype("datetime64[D]" )# mosaic along time dimension = flatten(sentinel_stack, dim= "time" )
/tmp/ipykernel_4612/1844268677.py:12: UserWarning: Converting non-nanosecond precision datetime values to nanosecond precision. This behavior can eventually be relaxed in xarray, as it is an artifact from pandas which is now beginning to support non-nanosecond precision values. This warning is caused by passing non-nanosecond np.datetime64 or np.timedelta64 values to the DataArray or Variable constructor; it can be silenced by converting the values to nanosecond precision ahead of time.
time=sentinel_stack.time.astype("datetime64[D]")
<xarray.DataArray 'stackstac-5efc1f91909c10ae7d8453451c8ac811' (time: 10,
band: 3,
y: 208, x: 325)> Size: 16MB
dask.array<concatenate, shape=(10, 3, 208, 325), dtype=float64, chunksize=(1, 1, 208, 325), chunktype=numpy.ndarray>
Coordinates: (12/21)
* band (band) <U5 60B 'red' 'green' 'blue'
* x (x) float64 3kB 3.74e+05 ... 4.0...
* y (y) float64 2kB 2.765e+06 ... 2....
mgrs:utm_zone int64 8B 15
s2:product_type <U7 28B 'S2MSI2A'
s2:processing_baseline <U5 20B '04.00'
... ...
raster:bands object 8B {'nodata': 0, 'data_ty...
common_name (band) <U5 60B 'red' 'green' 'blue'
center_wavelength (band) float64 24B 0.665 0.56 0.49
full_width_half_max (band) float64 24B 0.038 ... 0.098
epsg int64 8B 5070
* time (time) datetime64[ns] 80B 2022-0...
Attributes:
spec: RasterSpec(epsg=5070, bounds=(373900, 2744600, 406400, 27654...
crs: epsg:5070
transform: | 100.00, 0.00, 373900.00|\n| 0.00,-100.00, 2765400.00|\n| 0...
resolution: 100 dask.array<chunksize=(1, 1, 208, 325), meta=np.ndarray>
Bytes
15.47 MiB
528.12 kiB
Shape
(10, 3, 208, 325)
(1, 1, 208, 325)
Dask graph
30 chunks in 44 graph layers
Data type
float64 numpy.ndarray
10 1 325 208 3
Coordinates: (21)
band
(band)
<U5
'red' 'green' 'blue'
array(['red', 'green', 'blue'], dtype='<U5') x
(x)
float64
3.74e+05 3.74e+05 ... 4.064e+05
array([373950., 374050., 374150., ..., 406150., 406250., 406350.]) y
(y)
float64
2.765e+06 2.765e+06 ... 2.745e+06
array([2765350., 2765250., 2765150., ..., 2744850., 2744750., 2744650.]) mgrs:utm_zone
()
int64
15
s2:product_type
()
<U7
'S2MSI2A'
array('S2MSI2A', dtype='<U7') s2:processing_baseline
()
<U5
'04.00'
array('04.00', dtype='<U5') instruments
()
<U3
'msi'
array('msi', dtype='<U3') s2:datatake_type
()
<U8
'INS-NOBS'
array('INS-NOBS', dtype='<U8') s2:saturated_defective_pixel_percentage
()
int64
0
processing:software
()
object
{'sentinel2-to-stac': '0.1.0'}
array({'sentinel2-to-stac': '0.1.0'}, dtype=object) proj:epsg
()
int64
32615
constellation
()
<U10
'sentinel-2'
array('sentinel-2', dtype='<U10') title
(band)
<U20
'Red (band 4) - 10m' ... 'Blue (...
array(['Red (band 4) - 10m', 'Green (band 3) - 10m',
'Blue (band 2) - 10m'], dtype='<U20') proj:shape
()
object
{10980}
array({10980}, dtype=object) gsd
()
int64
10
raster:bands
()
object
{'nodata': 0, 'data_type': 'uint...
array({'nodata': 0, 'data_type': 'uint16', 'bits_per_sample': 15, 'spatial_resolution': 10, 'scale': 0.0001, 'offset': -0.1},
dtype=object) common_name
(band)
<U5
'red' 'green' 'blue'
array(['red', 'green', 'blue'], dtype='<U5') center_wavelength
(band)
float64
0.665 0.56 0.49
array([0.665, 0.56 , 0.49 ]) full_width_half_max
(band)
float64
0.038 0.045 0.098
array([0.038, 0.045, 0.098]) epsg
()
int64
5070
time
(time)
datetime64[ns]
2022-09-03 ... 2022-09-28
array(['2022-09-03T00:00:00.000000000', '2022-09-06T00:00:00.000000000',
'2022-09-08T00:00:00.000000000', '2022-09-13T00:00:00.000000000',
'2022-09-16T00:00:00.000000000', '2022-09-18T00:00:00.000000000',
'2022-09-21T00:00:00.000000000', '2022-09-23T00:00:00.000000000',
'2022-09-26T00:00:00.000000000', '2022-09-28T00:00:00.000000000'],
dtype='datetime64[ns]') Indexes: (4)
PandasIndex
PandasIndex(Index(['red', 'green', 'blue'], dtype='object', name='band')) PandasIndex
PandasIndex(Index([373950.0, 374050.0, 374150.0, 374250.0, 374350.0, 374450.0, 374550.0,
374650.0, 374750.0, 374850.0,
...
405450.0, 405550.0, 405650.0, 405750.0, 405850.0, 405950.0, 406050.0,
406150.0, 406250.0, 406350.0],
dtype='float64', name='x', length=325)) PandasIndex
PandasIndex(Index([2765350.0, 2765250.0, 2765150.0, 2765050.0, 2764950.0, 2764850.0,
2764750.0, 2764650.0, 2764550.0, 2764450.0,
...
2745550.0, 2745450.0, 2745350.0, 2745250.0, 2745150.0, 2745050.0,
2744950.0, 2744850.0, 2744750.0, 2744650.0],
dtype='float64', name='y', length=208)) PandasIndex
PandasIndex(DatetimeIndex(['2022-09-03', '2022-09-06', '2022-09-08', '2022-09-13',
'2022-09-16', '2022-09-18', '2022-09-21', '2022-09-23',
'2022-09-26', '2022-09-28'],
dtype='datetime64[ns]', name='time', freq=None)) Attributes: (4)
spec : RasterSpec(epsg=5070, bounds=(373900, 2744600, 406400, 2765400), resolutions_xy=(100, 100)) crs : epsg:5070 transform : | 100.00, 0.00, 373900.00|
| 0.00,-100.00, 2765400.00|
| 0.00, 0.00, 1.00| resolution : 100
Up until now, we have not processed any actual raster data! All of the operations have been carried out using the STAC item information and associated raster metadata from the source files. By working in this way, you can iterate very rapidly making sure that the dimensions of the output matches your expectation before you process any actual data.
Load the data into memory and take a look
You can keep going with an analysis 100% lazily until the very end, but this time I am just making a plot so we have reached the end of the lazy road. Calling the compute method will execute the lazily-evaluated operations that we queued up for flat_stack.
= flat_stack.compute()
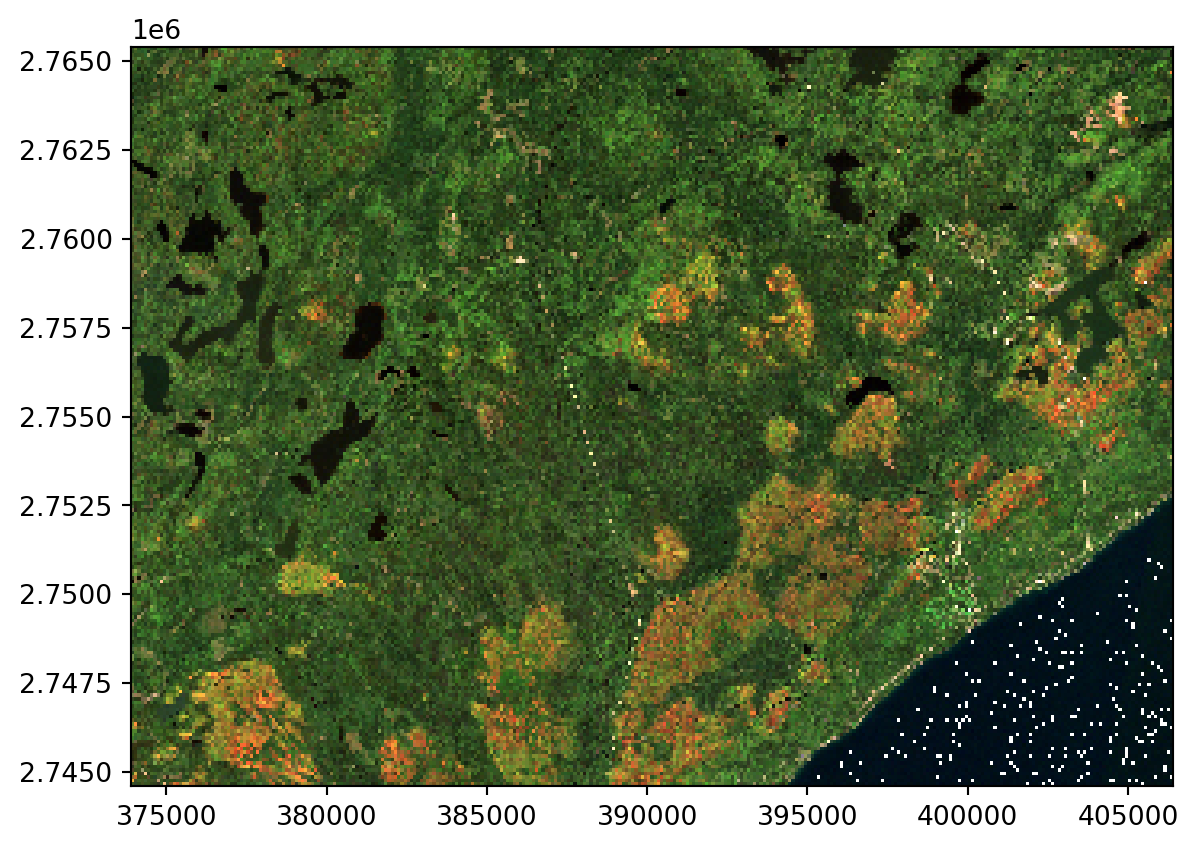
We can view the images easily with the plot method, giving us a glimpse into autumn on the North Shore of Lake Superior!
= BANDS).plot.imshow(= "time" ,= 4 ,= "band" ,= True ,= 4 ,=- 0.1 ,=- 0.01 ,= False ,
I remember it being cloudy last fall but damn we had a couple of nice days! Apparently September 28 was a really good one.
= BANDS, time= "2022-09-28" ).plot.imshow(= "band" ,= True ,= 5 ,=- 0.1 ,=- 0.01 ,= False ,
Whats next?
This is really just scratching the surface of what you can do with xarray, I will try to cover some more advanced topics in later posts.